|
Human Genetics
Inheritance, DNA and the Genetic Code |
|
|
|
Traditional Views of Heredity
It has been known since prehistoric times that the traits of parents are transmitted to offspring. Resemblance of siblings to one another and to their progenitors is a fact generally observed, as is the occurrence of variability in the inheritance of specific characteristics. For example, although the offspring of tall parents is often tall, this is not always so. Sometimes the son or daughter of tall parents is of average height, and sometimes the son or daughter of two brown-eyed parents has blue eyes. Nevertheless, the observed general tendency is that offspring inherit traits from both parents, although in a not totally predictable manner.
Close family members are known to share certain characteristics. Stature, facial features, hair color, and other characteristics are known to be often shared to some extent among siblings, cousins, and other close kin.
Around 420 B.C., the Greek physician Hippocrates of Kos theorized that kernels from various body parts of parents were transferred to their offspring. The traits were transmitted in some manner involving sexual intercourse.
The Discovery of Cellular Structure
 In 1665, Englishman Robert Hooke described in his book Micrographia his use of a microscope to view small objects. Micrographia had plates showing insects and miniature objects in a fine detail not published before. The book included a drawing of the structure of a slice of cork. Hooke discovered cork had an internal structure made up of what looked like miniature rooms. The word cell, which Hooke was the first to use in this context, derives from the Latin cella (small room).
In 1665, Englishman Robert Hooke described in his book Micrographia his use of a microscope to view small objects. Micrographia had plates showing insects and miniature objects in a fine detail not published before. The book included a drawing of the structure of a slice of cork. Hooke discovered cork had an internal structure made up of what looked like miniature rooms. The word cell, which Hooke was the first to use in this context, derives from the Latin cella (small room).
The roughly oblong features that Hooke described were the walls of dead tree cells. The existence of microscopic living creatures (microorganisms) was documented by Anton van Leeuwenhoek in 1675. Most cells are between 1/250-inch and 1/2500-inch in size, so it was only after the invention of the microscope that they became visible. Van Leeuwenhoek, a Dutch fabric merchant and lens maker, constructed small single-lens microscopes capable of magnifying objects over 275 times. In letters to the Royal Society of London, van Leeuwenhoek contributed for publication numerous illustrations of his observations of microorganisms (bacteria, protists, and animal and plant cells).
The growing availability of microscopes led to further discoveries and it gradually became apparent that there were many types of single-cell organisms and that plant and animal tissues are comprised of cells. By 1839, Germans Matthias Schleiden and Theodor Schwann had developed a cell theory applying to plant and animal tissues. Schwann wrote in Microscopic Investigations on the Accordance in the Structure and Growth of Plants and Animals that all living things are composed of cells and cell products.
Origin of Microorganisms
French chemist Louis Pasteur conducted experiments which demonstrated that fermentation is caused by the growth of microorganisms. In his 1857 paper, Report on the Lactic Acid Fermentation, Pasteur identified an agent, which he called lactic yeast, as the cause of the production of lactic acid from sugar. Lactic yeast, the lactic acid bacteria group, make lactic acid as a metabolic product of carbohydrate fermentation. Pasteur went on to disprove the concept of spontaneous generation, and to stablish in 1862 that microorganisms come to be only from parents similar to themselves.
Pangenesis
The understanding of cellular structure led Charles Darwin to build on the earlier inheritance theory of Hippocrates and on his own theory of sexual selection. In his 1868 book, The Variation of Plants and Animals Under Domestication, Darwin theorized that general body cells in living things transfer minute granules (via the bloodstream in animals) to the reproductive organs.
This theory, Pangenesis, hypothesized that the granules, called gemmules, were the basic units of hereditary transmission. The gemmules were incorporated in the reproductive cells, so general body cells were represented in the germ cells that eventually determined the makeup of the offspring through a form of blending inheritance.
Gregor Mendel and the Discovery of Hereditary Factors
 Gregor Mendel was an Austrian physicist and Augustinian friar who conducted a series of experiments in inheritance from 1856 to 1863. He was painstakingly rigorous in his methodology and record keeping, and used very large samples and sound statistical techniques. In 1866, Mendel published his paper, Experiments on Plant Hybridization, in which he described what are now known as Mendelís Laws of Inheritance.
Gregor Mendel was an Austrian physicist and Augustinian friar who conducted a series of experiments in inheritance from 1856 to 1863. He was painstakingly rigorous in his methodology and record keeping, and used very large samples and sound statistical techniques. In 1866, Mendel published his paper, Experiments on Plant Hybridization, in which he described what are now known as Mendelís Laws of Inheritance.
Despite the initial unpredictability of his observed results, Mendel was able to arrive at his conclusions about inheritance because of his insight and the large scope of his research. Mendel used 22 varieties of garden pea in his hybridization experiments, and observed the expression of seven different characteristics. Analysis of the massive volume of data he collected led him to infer patterns in the inheritance of traits and to confirm his hypotheses through further experiments. He discovered that certain traits were dominant and others were recessive. For example, the round seed shape characteristic was dominant, the wrinkled seed shape was recessive.
Mendel devised the concept of hereditary factors, one a recessive characteristic and the other a dominant characteristic. Mendel found that each individual has two factors for each characteristic, one from each parent. The two factors may be the same or may be different. If the two factors are identical, the individual is called homozygous for the characteristic. If the two factors are different, the individual is heterozygous for that characteristic.
Designating the dominant form of the factor A, and the recessive a, three possible pairings may be encountered: AA, aa, and Aa. The three alternative types of the factor are called alleles. Mendelís experiments showed that the Aa allele, the hybrid form, occurred approximately twice as often as the AA and aa forms, since it can be produced in two different ways: Aa and aA. The dominant characteristic is expressed when the AA or the Aa allele is inherited. (The dominant A form masks the recessive form a when Aa is inherited.) The recessive characteristic only manifests itself when the aa allele is inherited.
The First Mendel Law, the Law of Segregation, states that every individual possesses a pair of alleles for any particular characteristic and that each parent passes one randomly selected allele to its offspring. For each characteristic, the offspring receives one allele from each parent, resulting in its own pair of alleles. Whether the offspring expresses the dominant or recessive form of the characteristic depends on the inherited allele pair.
Mendelís Second Law, the Law of Independent Assortment, states that separate alleles for separate characteristics are transmitted from parents to offspring independently of one another. The selection of a particular allele in the allele pair for one characteristic transmitted to the offspring is not related to the selection of an allele for other characteristics. (Mendel concluded that there is no relation, for example, between a garden pea plantís shape and its flower color.)
Mendelís observations explained why a parentís characteristic can sometimes recur in offspring and other times disappear, depending on the inheritance of either the dominant (AA), recessive (aa), or hybrid (Aa) allele of a specific characteristic. The concept of alleles also explained the reappearance of a previously repressed characteristic. An individual with an Aa allele would not display the recessive form of the factor, but two individuals with the Aa allele, who would both display the dominant A characteristic, could produce offspring displaying the recessive characteristic (in this case, statistically, the aa allele would appear 25 percent of the time). Two individuals with the aa allele would both display the recessive characteristic and would transmit it to their offspring through their union.
Mendelís work provided the basis for the understanding of inheritance as a discontinuous rather than a blending process.
Chemical Composition and Structure of the Cell Nucleus
 By the mid-nineteenth century, the basic elements of cells were identified: an outer wall, a cytoplasm, and a nucleus. Within the cytoplasm are a number of structures, among them mitochondria, vacuoles, the Golgi apparatus, and ribosomes. The internal structure of the nucleus gradually became known.
By the mid-nineteenth century, the basic elements of cells were identified: an outer wall, a cytoplasm, and a nucleus. Within the cytoplasm are a number of structures, among them mitochondria, vacuoles, the Golgi apparatus, and ribosomes. The internal structure of the nucleus gradually became known.
A few years after Mendelís experiments, Swiss physician Johannes Friedrich Miescherís research on the chemical composition of cell nuclei led him to identify chemical compounds that he called nuclein. In 1869, Miescher, working at the University of Tubingen in Germany, was able to extract nuclein from the nuclei of human white cells (leucocytes). Miescher determined that nuclein contained Nitrogen, Phosphorous, Oxygen, and Hydrogen. He observed the curious fact that, in nuclein, Phosphorous and Nitrogen occurred in a fixed ratio.
German biochemist Ludwig Albrecht Kossel further analyzed the chemical composition of nuclein. Kossel determined that nuclein was constituted of organic (carbon-based) compounds, nucleic acids. In the period from 1885 to 1901, Kossel identified the nucleic acids adenine, cytosine, guanine, thymine, and uracil.
 Walther Fleming, a German biologist, discovered the presence of threadlike structures in cell nuclei. Fleming used aniline dyes to visualize intracellular structures with his microscope. He named the threadlike structures chromosomes (colored bodies). Fleming studied cell division, a process he called mitosis, and in 1882 published Cell Substance, Nucleus and Cell Division, concluding that cell nuclei derive from predecessor cell nuclei, with chromosomes being distributed from parent nuclei to descendant nuclei.
Walther Fleming, a German biologist, discovered the presence of threadlike structures in cell nuclei. Fleming used aniline dyes to visualize intracellular structures with his microscope. He named the threadlike structures chromosomes (colored bodies). Fleming studied cell division, a process he called mitosis, and in 1882 published Cell Substance, Nucleus and Cell Division, concluding that cell nuclei derive from predecessor cell nuclei, with chromosomes being distributed from parent nuclei to descendant nuclei.
In 1883, Belgian zoologist Edouard Van Beneden described the role of chromosomes in a type of cell division called meiosis. Unlike in mitosis, where cell division results in two cells with identical nuclear material, in meiosis chromosomes undergo a recombination, and cell division results in gametes (ova in females and spermatozoa in males) which are not identical. Seven years later, in 1890, German biologist August Weismann provided a more detailed explanation of meiosis, noting that in meiosis two cell divisions are needed to convert one diploid cell (containing two chromosome strands) into four haploid cells (each with one unique chromosome strand).
Dissemination of Mendelís Theories
German botanist Carl Erich Correns began to study plant trait inheritance in 1892. In 1900 he published the paper Georg Mendelís Law Concerning the Behavior of the Progeny of Racial Hybrids, in which he presented his own results, restated Mendelís principles of inheritance, and provided support for Mendelís theories, which had not been widely circulated. At about the same time, Dutch botanist Hugo de Vries and Austrian agronomist Erich von Tschermak-Seysenegg independently arrived at similar conclusions. De Vries theorized that inheritance of specific traits is carried in particles he called pangenes.
The Physical Basis of Heredity
 American physician Walter Sutton discovered through his work on grasshopper chromosomes that the nuclein filaments occur in pairs within the nucleus and separate during meiosis, providing a vehicle for cells, and therefore male and female parents, to pass on their genetic material. In 1903, Sutton published The Chromosomes in Heredity, which presented the theory that the association of paternal and maternal chromosomes in pairs and their subsequent separation constituted the physical basis of the Mendelian laws of heredity.
American physician Walter Sutton discovered through his work on grasshopper chromosomes that the nuclein filaments occur in pairs within the nucleus and separate during meiosis, providing a vehicle for cells, and therefore male and female parents, to pass on their genetic material. In 1903, Sutton published The Chromosomes in Heredity, which presented the theory that the association of paternal and maternal chromosomes in pairs and their subsequent separation constituted the physical basis of the Mendelian laws of heredity.
Genetics
English scientist William Bateson studied structural variations in living organisms. Bateson observed the occurrence of hairy and smooth forms of certain plants, without intermediate types. In 1894 he wrote in Materials for the Study of Variation that biological variation occurs in two forms, a continuous form showing a blending of traits, and a discontinuous type where different varieties of a trait were encountered and intermediate forms were absent. Speaking in 1906 at the Third International Conference on Plant Hybridization, Bateson first used the word genetics to refer to the study of inheritance and heredity.
In 1909, in his book Elements of the Exact Heredity, Danish botanist Wilhelm Johannsen introduced the term gene to refer to particles carrying inheritable traits. Johannsen made the distinction between an organismís observable characteristics, which he called the phenotype (shown type), and the inherited characteristics, which he called the genotype. Johannsen postulated that an organismís observable characteristics are the result of inheritance, but also of environmental factors. For example, a plantís height is determined by its inherited characteristics, but also by water and nutrients available during its development.
Hans von Miniwarter, an Austrian-born Belgian cytologist, was able to visualize individual human chromosomes during cell division. In 1912, he estimated (incorrectly) the total number of chromosomes as 47 in sperm cells and 48 in ova. Theophilus S. Painter, a zoologist at the University of Texas, estimated in 1921 the number of chromosomes in humans as 46, 47, or 48. Painter correctly identified chromosome X as the female sex chromosome and Y as the male sex chromosome.
American biologist Thomas Hunt Morgan conducted hereditary experiments using the fly Drosophila melanogaster. Morgan, then at Columbia University, concluded that chromosomes carry genetic information in meiosis, and are the physical basis of heredity. In 1915, Morgan, Alfred Sturtevant, Calvin Bridges and Hermann Muller published The Mechanism of Mendelian Heredity, presenting experimental support for Suttonís theory and affirming that chromosomes carry genes and constitute the physical basis for heredity.
Nucleotides
After 1915, a consensus gradually developed that a gene is the unit of heredity (in the sense of Mendelís hereditary factors), a chromosome includes a set of genes, and nucleic acids in the cell nucleus incorporate an assemblage of chromosomes. Although microscopes made it possible to visualize chromosomes during certain phases of cell division, genes remained too small to be seen and their exact nature was unknown. The chemical components of nucleic acids were known: Carbon, Nitrogen, Hydrogen, Oxygen, and Phosphorous. But the molecular structure of nucleic acids was largely a mystery, although it was realized that there are two types, deoxyribonucleic acid and ribonucleic acid, both polymers. Polymers are chemical compounds comprising repeating structural units linked by chemical bonds.
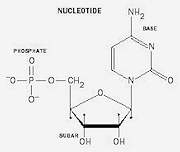 By 1929, Lithuanian-American biochemist Phoebus Levene, working at the Rockefeller Institute of Medical Research, had figured out that nucleic acid polymers are made up of nucleotides. Nucleotides have three component units, a five-carbon sugar, a phosphate group, and a nitrogenous base.
By 1929, Lithuanian-American biochemist Phoebus Levene, working at the Rockefeller Institute of Medical Research, had figured out that nucleic acid polymers are made up of nucleotides. Nucleotides have three component units, a five-carbon sugar, a phosphate group, and a nitrogenous base.
Jean Louis Brachet, a Belgian biochemist, discovered in 1933 that deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) occur in both animal and plant cells. Brachet found that chromosomes contain DNA, and that RNA occurs in the cytoplasm. The cytoplasm includes components enveloped by the cell membrane but outside the nucleus, such as organelles and cytoplasmic inclusions.
Brachet noted that RNA was involved in protein synthesis within cells. He demonstrated experimentally (by removing the nucleus of cells and observing the gradual cessation of protein synthesis) that RNA is produced in the cell nucleus and is then conveyed to the cytoplasm.
Molecular Structure and X-ray Crystallography
Born in New Zealand, but doing much of his work in England, Ernest Rutherford, a physicist and chemist, developed a theory of atomic structure. In 1910 he proposed that atoms consist of a small, relatively heavy, positively charged nucleus, with much lighter negatively charged electrons in orbit around it. The nucleus contains neutrons, which have no charge, and positively charged protons. In 1913, Danish Physicist Niels Bohr proposed that electrons travel only in certain successively larger orbits around the atomic nucleus. The outer orbits can hold more electrons than the inner ones, and determine the atomís chemical properties.
Molecules are groups of two or more atoms held together by covalent bonds. Living organisms incorporate complex group molecules. Some are polymers, including peptides, the nucleic acids, and polysaccharides. Other group molecules include amino acids (monomer building blocks of proteins and peptides) and the lipids.
 Crystals are composed of periodic arrays of atoms. When exposed to X-rays, crystals cause distinct diffraction patterns due to their internal molecular structure. In 1912, British physicist William Lawrence Bragg developed a method to determine the positions of the atoms within a crystal from the way in which an X-ray beam is diffracted by the crystal lattice. Bragg developed X-ray crystallography as a method for determining the geometry of organic molecules.
Crystals are composed of periodic arrays of atoms. When exposed to X-rays, crystals cause distinct diffraction patterns due to their internal molecular structure. In 1912, British physicist William Lawrence Bragg developed a method to determine the positions of the atoms within a crystal from the way in which an X-ray beam is diffracted by the crystal lattice. Bragg developed X-ray crystallography as a method for determining the geometry of organic molecules.
In 1948, working at the University of Cambridge Cavendish Laboratory, Bragg led work on the structure of proteins. Using crystallography, scientists at Cavendish Laboratory were analyzing by 1950 the 3-dimensional structure of polypeptide molecules. These molecules have the shape of a cylindrical helix. American chemist Linus Pauling used X-ray crystallography images to study protein molecular structure in 1951.
In 1952, Linus Pauling and Robert Corey, at Caltech, analyzed X-ray crystallography images of nucleic acids, the work of William Astbury, to resolve the molecular structure of DNA. Pauling developed a concept involving a three-chain molecule, with each chain being a helix with the bases facing outward and the phosphates in the center. The resulting model approximately fit the crystallography data. Pauling and Corey published their paper, A Proposed Structure for the Nucleic Acids, in the Proceedings of the Natural Academy of Sciences on February 1953. Paulingís proposed structure for DNA was deeply flawed. It was soon determined that the proposed structure did not provide enough space at the center to accommodate all the phosphates, and the deviation from crystallography data was excessive. The three-chain concept was deemed unsatisfactory.
The Molecular Structure of DNA
In the United Kingdom, Biophysicists Rosalind Elsie Franklin and Maurice Wilkins used X-ray crystallography in the period from 1951 to 1953 to study the structure of DNA molecules at Kingís College London. At the Cavendish Laboratory, American zoologist James Watson and British physicist Francis Crick analyzed the structure of DNA. Their approach was to build physical molecular models, guided by available X-ray crystallography images, to evaluate possible chemical arrangements and incrementally arrive at an accurate representation of the DNA molecule.
 Early in 1953, Watson and Crick completed a detailed 3-dimensional model of the deoxyribonucleic acid molecule that perfectly fit data obtained from X-ray crystallography images. The Watson-Crick stick-and-ball model showed the DNA molecule to have the shape of a double helix, with two chains, going in reverse directions. Matching base pairs interlocked at the center of the double helix, keeping constant the distance between the chains. The bases were on the inside, and the phosphate backbone was on the outside of the cylinder formed by the double helix.
Early in 1953, Watson and Crick completed a detailed 3-dimensional model of the deoxyribonucleic acid molecule that perfectly fit data obtained from X-ray crystallography images. The Watson-Crick stick-and-ball model showed the DNA molecule to have the shape of a double helix, with two chains, going in reverse directions. Matching base pairs interlocked at the center of the double helix, keeping constant the distance between the chains. The bases were on the inside, and the phosphate backbone was on the outside of the cylinder formed by the double helix.
 William Bragg, Director of the Cavendish Laboratory, first announced Watson and Crickís discovery of the molecular structure of DNA at the Solvay Conference on Proteins in Brussels on April 8, 1953. Watson and Crick published their paper, A Structure for Deoxyribose Nucleic Acid in the journal Nature on April 25, 1953. In the same issue of Nature, Maurice Wilkins, A. Stokes and H. Wilson, and Rosalind Franklin and Ray Gosling published companion papers describing supporting X-ray crystallography evidence. On May 30, 1953, Watson and Crick published a follow-up paper in Nature, Genetical Implications of the Structure of Deoxyribonucleic Acid, describing how base-pairing supports DNA replication.
William Bragg, Director of the Cavendish Laboratory, first announced Watson and Crickís discovery of the molecular structure of DNA at the Solvay Conference on Proteins in Brussels on April 8, 1953. Watson and Crick published their paper, A Structure for Deoxyribose Nucleic Acid in the journal Nature on April 25, 1953. In the same issue of Nature, Maurice Wilkins, A. Stokes and H. Wilson, and Rosalind Franklin and Ray Gosling published companion papers describing supporting X-ray crystallography evidence. On May 30, 1953, Watson and Crick published a follow-up paper in Nature, Genetical Implications of the Structure of Deoxyribonucleic Acid, describing how base-pairing supports DNA replication.
DNA is made up of paired nucleotide units. Hydrogen bonds hold together chains of guanine-cytosine and adenine-thymine base pairs. The length of the guanine-cytosine and the adenine-thymine base pair is the same, so they fit evenly between the two phosphate backbones. The planes of the bases are perpendicular to the fiber axis. The hydrogen bonds are easily broken and reformed, providing a way to unzip the two complementary DNA strands, facilitating copying the genetic material. Since only specific pairs of bases can be formed, if the sequence of bases on one chain is given, then the sequence on the other chain is automatically determined.
Number of Human Chromosomes
 Although human chromosomes were individually discerned via microscopes since 1912, it was not until the middle of the twentieth century that the exact number was determined with certainty. The difficulty in visualization stemmed from the 3-dimensional arrangement of chromosomes within cells, from the need to see the nuclear material part-way through cell division, and from available chromosome staining and placement techniques. In 1956, two independent papers (C.E. Ford and J.L. Hamerton, The Chromosomes of Man, Nature 178, and Joe Hin Tjio and Albert Levan, The Chromosome Number of Man, Hereditas 42) were published establishing the number of chromosomes in human somatic cells as 23 pairs (46 total).
Although human chromosomes were individually discerned via microscopes since 1912, it was not until the middle of the twentieth century that the exact number was determined with certainty. The difficulty in visualization stemmed from the 3-dimensional arrangement of chromosomes within cells, from the need to see the nuclear material part-way through cell division, and from available chromosome staining and placement techniques. In 1956, two independent papers (C.E. Ford and J.L. Hamerton, The Chromosomes of Man, Nature 178, and Joe Hin Tjio and Albert Levan, The Chromosome Number of Man, Hereditas 42) were published establishing the number of chromosomes in human somatic cells as 23 pairs (46 total).
The term karyotype denotes the set of chromosomes in the nucleus of a eukaryotic cell. In human diploid cells, chromosomes occur in twenty-two autosomes, chromosome pairs that have the same form, and one allosome (sex chromosome) pair that consists of two X chromosomes (female) or one X chromosome and one Y chromosome (male).
Decoding the DNA Molecule
The DNA molecule stores information using a four-letter alphabet consisting of the nitrogenous bases adenine (A), guanine (G), thymine (T), and cytosine (C). Its great size (each strand of a human DNA molecule, if uncoiled and laid out linearly, is about 40 inches long) makes it evident that the information occurs in vast amounts. Once the general structure of the DNA molecule became known, speculation turned to discerning the uses the information was put to, and the chemical mechanisms involved.
Three basic uses of DNA were apparent: the replication of the cell itself via mitosis; in cases involving meiosis and the reproductive organs, the procreation of the individual organism; and the production of the proteins needed to support life.
The Central Dogma of Molecular Biology
 In 1956, Francis Crick drafted a paper in which he formulated general rules for detailed sequence information transfer from one polymer with a defined alphabet to another. Crick presented the Central Dogma of Molecular Biology in the paper Ideas on Protein Synthesis at the Symposium of the Society of Experimental Biology in 1958.
In 1956, Francis Crick drafted a paper in which he formulated general rules for detailed sequence information transfer from one polymer with a defined alphabet to another. Crick presented the Central Dogma of Molecular Biology in the paper Ideas on Protein Synthesis at the Symposium of the Society of Experimental Biology in 1958.
In its most basic form, the Central Dogma states that once information has got into a protein, it cannot get out again. The principle is that DNA can make DNA, DNA can make RNA, and RNA can make protein, but protein cannot make RNA, DNA, or another protein. Only under special circumstances can RNA make RNA or DNA. Basically, the nucleotide units of nucleic acids and requisite enzymes provide a complex molecular mechanism that works to assemble proteins, and there is no known method for proteins to reverse or replicate the process. Crick went on to publish a more rigorous version of this principle in his paper, Central Dogma of Molecular Biology, in the August 1970 Nature.
Production of Proteins Within the Cell
In 1959, Spanish-born American biochemist Severo Ochoa received the Nobel Prize for Physiology or Medicine for discovery of an enzyme, polynucleotide phosphorylase, that allowed him to synthesize RNA. Ochoa, at New York University Medical School, conducted research to find out how RNA is involved in making proteins in the cell.
The means by which DNA directs the making of proteins began to be elucidated in 1961, when Marshall Nirenberg, an American biochemist, presented a paper at the International Congress of Biochemistry in Moscow indicating that messenger RNA (mRNA) directs protein synthesis. Working with Johann Heinrich Matthaei, Nirenberg, then with the National Institutes of Health, had conducted experiments using the bacterium Escherichia coli and a synthetic RNA molecule which led to the discovery that a three-nucleotide sequence of uracil coded for the amino acid phenylalanine. In August 1961, Nirenberg and Matthaei published their paper, The Dependence of Cell-Free Protein Synthesis in E. Coli upon Naturally Occurring or Synthetic Polyribonucleotides, in the Proceedings of the National Academy of Sciences.
The Nirenberg-Matthaei experiment was an initial step in the solution of the code used in the production of proteins. Har Khorana, a Pakistani-American biologist, worked out the three-nucleotide sequences (codons) that coded for the amino acids serine and leucine. Khoranaís experiments also led to finding that certain codons in RNA (stop codons) signal the termination of the process that translates mRNA to produce proteins. In 1964, Robert Holley, an organic chemist then with Cornell University, worked out the chemical sequence and structure of the molecule that incorporates the amino acid alanine into proteins. The discovery of this molecule, alanine transfer RNA (tRNA), was a key finding in explaining the production of proteins from messenger RNA. In 1966, Holley published the results of his work in The Nucleotide Sequence of a Nucleic Acid.
Once the first codons were deciphered, there was a concerted effort to discover the RNA codes for the major amino acids. With this knowledge, it would be possible to not only know how RNA works to translate messages from DNA to build proteins, but to read the complete genetic code of living organisms. Soon the three-letter RNA codes (codons) for each of the 20 amino acids common in living organisms were determined. There are hundreds of other amino acids that appear in nature, but not in living organisms. All 64 possible three-letter combinations have a corresponding amino acid, or code for a stop codon. In most cases, several different codons can code for the same amino acid.
The production of proteins within the cell involves two distinct processes. The first, transcription, takes place within the cell nucleus and directly concerns DNA and RNA. The second process, translation, takes place outside the nucleus in cellular organelles called ribosomes and involves RNA.
Unlike DNA, where the two strands are arranged in a double helix, the RNA molecules consist of a single non-helical strand. RNA contains ribose sugar molecules, rather than the deoxyribose molecules in DNA.
Transcription
The transcription process uses a part of the DNA molecule as a template for making an RNA molecule. A helicase enzyme splits the DNA molecule into two nucleotide strands. The process advances when the enzyme RNA polymerase binds to a DNA strand. The enzyme makes an RNA strand corresponding to a specific stretch of DNA by assembling a chain of nucleotides. Resulting complementary RNA nucleotides are arranged in reverse order to the nucleotide sequence in the DNA template.
DNA is made up of the bases adenine, cytosine, guanine, and thymine. RNA is similar, but differs in that it does not incorporate thymine. In RNA, the nitrogenous base uracil replaces thymine as the complementary nucleotide to adenine. The RNA molecule stores information using a four-letter alphabet consisting of the nitrogenous bases adenine (A), guanine (G), uracil (U), and cytosine (C). During transcription, when the RNA polymerase encounters adenine in the DNA strand, it attaches a uracil nucleotide in the RNA strand. Uracil in RNA is informationally analogous to thymine in DNA, thus giving four standard symbols for the components of nucleic acid.
The transcription process continues producing an increasingly longer strand of RNA until a termination indicator is encountered. In eukaryotes (organisms whose cells contain a nucleus) this happens when a specific series of nucleotides appears along the DNA strand. In some cases, a protein known as a termination factor is also necessary for transcription to be concluded.
When the mRNA molecule is terminated, it separates from the DNA template. In eukaryotes, the mRNA then undergoes an editing process. Sections of the mRNA made up of nucleotide sequences that do not code for proteins are often cropped off. These intervening nucleotide sequences are called introns. After the introns are removed, a short sequence of adenine nucleotides is added to the end of the mRNA molecule. This set of nucleotides, called a poly-A tail, marks the mRNA molecule, which can then travel across the wall of the nucleus into the cytoplasm.
Translation
Translation produces proteins. It takes place in the ribosomes and involves the molecules messenger RNA (mRNA), transfer RNA (tRNA), and ribosomal RNA (rRNA).
During translation, the mRNA transported to the cytoplasm from the nucleus is decoded to produce the precise sequence of amino acids in a protein. Translation requires numerous enzymes.
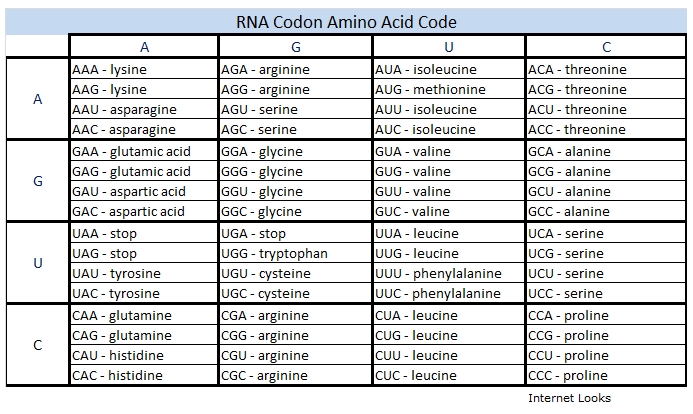
Protein sequences are translated beginning at a place defined by a start codon, usually the first AUG codon in the mRNA codon series. The codons UUG and GUG can in some cases also be used as start codons. It is critical that the beginning of translation be precisely identified, so the start of translation requires the start codon plus nearby sequences and initiation factors. Start codons are translated as the amino acid methionine (formylmethionine in bacteria). When not functioning as start codons, UUG codes for leucine and GUG for valine.
Ribosomes are organelles formed by ribosomal RNA (rRNA) molecules associated with other proteins. Transfer RNA (tRNA) molecules are small RNA molecules that carry an anticodon region and a specific associated amino acid.
Translation is initiated in the cytoplasm when an mRNA molecule becomes associated with ribosomes. Transfer RNA molecules in the ribosomes, each carrying a specific amino acid, link with codons in the mRNA. Each tRNA anticodon binds with a corresponding mRNA codon. Since adenine (A) pairs with uracil (U), and guanine (G) pairs with cytosine (C), each three-letter mRNA codon has a corresponding tRNA anticodon and amino acid.
All protein coding sequences start with the initiation codon AUG. Also, all protein coding sequences end with a stop codon (UAA, UGA, or UAG). As the ribosome moves along the mRNA, the tRNA transfers its amino acid to the growing protein chain, producing the protein codon by codon. Protein translation stops when the ribosome encounters a stop codon.
Each chromosome includes a single DNA molecule. Genes are DNA segments arranged sequentially along DNA molecules. Each gene contains instructions for assembling a specific RNA or protein molecule.
Some portions of DNA molecules may not code for genes. The functions of those DNA portions are not well understood. Some appear to be copies of DNA segments, and may contain information useful in DNA repair.
Microorganisms
Microorganisms, living entities typically too small to be seen without microscopes, include archaea, bacteria, eukaryotes, and viruses. These smallest lifeforms exist in soil, subsurface, rivers, oceans and air. Some dwell on the skin and gut of humans. The total mass of all microorganisms on Earth exceeds the combined mass of all plants and animals.
Archaea and bacteria are classified as prokaryotes and lack a nucleus or membrane-bound organelles. In prokaryotes, DNA and other cell components are enclosed together within the outer cell membrane. Eukaryotes have a nucleus and typically are larger than prokaryotes; they include protozoa, algae, fungi, and slime molds. Individual cells of plants and animals are eukaryotic.
Viruses lack a cellular structure and can replicate only inside the cells of prokaryotes or eukaryotes; they consist of DNA or RNA, a coat of protein, and in some cases a coat of lipids. Extremely small, viruses are mostly too small to be seen with optical microscopes but can be observed using electron microscopes.
Because the genome of microorganisms is correspondingly small, the first efforts at genome sequencing were directed at microorganisms.
Genome Sequencing
The decoding of DNA and RNA and the understanding of the role of nucleic acids and enzymes in protein synthesis and in cellular and organism reproduction led, in the later part of the twentieth century, to efforts to develop entire genetic sequences for living organisms.
In 1995, a team led by John Craig Venter, an American biochemist and physiologist, succeeded in sequencing the entire genome of an organism, Haemophilus influenzae. This bacterium causes various diseases in humans (but not influenza, which is caused by a virus). Venter used a method called shotgun sequencing, in which DNA molecules are first divided randomly into segments which are independently sequenced and later reassembled. Haemophilus influenzae DNA was found to consist of 1.8 million base pairs, and contains 1,700 genes. The bacteriumís genome was described by R.D. Fleischmann et al. in their paper Whole-genome Random Sequencing and Assembly of Haemophilus influenzae Rd, in Science, Vol. 269. The genome revealed unexpected features, including repeated sequences, insertions, and inversions.
The complete genome sequence of the archaeon Methanococcus jannaschii (a primitive single-cell organism without a cell nucleus) was described in 1996 by C.J. Bult et al., in Science, Vol. 273. Sequencing methodology improved rapidly, and the genomes of various other organisms were soon worked out. In 2000, the complete genome of the fly Drosophila melanogaster was sequenced. D. melanogasterís DNA consists of 120 million base pairs and has 13,600 genes.
 In 2001, John Craig Venter and others, associated with Celera Genomics, published The Sequence of the Human Genome in Science, Vol. 291. Celera arrived at the genome sequence using a method called whole genome shotgun sequencing. The reference DNA sequence combined results obtained from the DNA of several individuals. The human genome has about three billion DNA base pairs and 25,000 genes.
In 2001, John Craig Venter and others, associated with Celera Genomics, published The Sequence of the Human Genome in Science, Vol. 291. Celera arrived at the genome sequence using a method called whole genome shotgun sequencing. The reference DNA sequence combined results obtained from the DNA of several individuals. The human genome has about three billion DNA base pairs and 25,000 genes.
The Human Genome Project, an international research project led by the United States Department of Energy and the National Institutes of Health, announced in 2003 the sequencing of a complete human genome. The Human Genome Project used a method called hierarchical shotgun sequencing, which divided the DNA molecule into sections that were mapped to chromosomes before being sequenced.
The genomes of various plants and vertebrates have been sequenced. They show a wide range of diversity, and of common features, including repeated sequences, insertions, and inversions. The genome of corn, for example, with 2.3 billion base pairs and about 32,000 genes, is comparable in size to the human genome.
The closer organisms are in terms of their species relationship, the greater their degree of genome commonality. But even distantly related organisms, such as horses and salamanders, share some genes. This is because genes code for many proteins that are common to most organisms, and many organisms share a similar general body plan or have organs that perform similar functions. Horses and salamanders both have a mouth, a brain, two eyes, a stomach, four limbs, and a tail, for example.
Human Genome Characteristics
The cells in the human body, except for reproductive cells, have 23 pairs of chromosomes in the nucleus (a total of 46 chromosomes). These somatic (body) cells are diploid, that is, they have two homologous copies of each chromosome, one from the mother and one from the father. The term diploid derives from the Greek diploos (double).
Human sex cells (gametes) have one set of 23 chromosomes, with one of the chromosomes being a sex chromosome, either male or female. These reproductive cells (sperm in males, ova in females) are haploid; they have only one copy of each chromosome. The word haploid derives from the Greek haploos (single).
DNA in the cell nucleus contains genes, which carry information for making proteins, as well as other nucleotide sequences which help regulate gene activity. Genes make up only about two percent of nuclear DNA; much of the rest is involved in producing RNA with gene regulating functions. Gene regulators are responsible for cell differentiation. All body cells of a particular individual have the same set of genes, but the genes operate differently in different cell types. For example, gene regulators may cause a set of genes to be active in kidney cells, and another set in heart cells.
The mitochondrion, a cellular organelle, contains its own DNA organized as several copies of a single circular chromosome. Mitochondria are involved in energy production and regulation of cell metabolism. Human mitochondrial DNA consists of a circular chromosome with 16,000 base pairs and 37 genes. The mitochondrial DNA code used in the production of proteins is in some respects different from the nuclear DNA code. In human mitochondria, the codons AGA and AGG are stop codes, AUA, AUC, and AUU are start codes, AUA codes for methionine, and UGA codes for tryptophan. In humans and most organisms, mitochondria are typically inherited only from the female parent.
In humans and other species that produce two distinct types of gametes, the male produces a mobile elongated sex cell (sperm), and the female a much larger round cell (ovum). Each gamete contributes half of the genetic information for the offspring. Fertilization results in a diploid cell, a zygote, which contains the chromosome sets of the sperm and the ovum. The sex of the offspring is determined by the sperm. If its sex chromosome is X, the child inherits two X chromosomes (ova always carry an X chromosome) and is female. If the sperm cell carries a Y sex chromosome, the child inherits one X and one Y chromosome and is male.
Chromosome Structure and Function
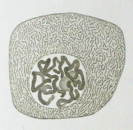
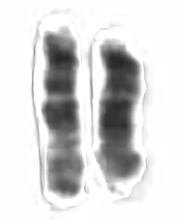 Each human chromosome has two strands of DNA, one inherited from the father, the other from the mother. In addition to holding the genetic code, chromosomes hold proteins useful in gene expression. Chromosomes display a banded structure, as shown in a micrograph of chromosome 15. Chromosome 15 has about 100 million DNA building blocks (base pairs). It contains approximately 570 protein-coding genes. One of the genes present in chromosome 15, PDCD7, encodes for the Programmed Cell Death 7 protein. Another gene, SLC24A5, codes for about a third of skin color differences between races.
Each human chromosome has two strands of DNA, one inherited from the father, the other from the mother. In addition to holding the genetic code, chromosomes hold proteins useful in gene expression. Chromosomes display a banded structure, as shown in a micrograph of chromosome 15. Chromosome 15 has about 100 million DNA building blocks (base pairs). It contains approximately 570 protein-coding genes. One of the genes present in chromosome 15, PDCD7, encodes for the Programmed Cell Death 7 protein. Another gene, SLC24A5, codes for about a third of skin color differences between races.
Gestation
The zygote spends a few days traveling down the motherís Fallopian tube, dividing to form a globe of cells, the morula. The zygote continues to divide, creating an inner cluster of cells surrounded by an outer shell. This stage of development is called a blastocyst. The inner cluster of cells eventually become the embryo, and the outer shell becomes the membranes that nourish and protect the embryo. The blastocyst reaches the uterus about five days after conception, and implants into the wall of the uterus around day six. By this stage in the motherís menstrual cycle, the lining of the uterus has grown and is ready to support a fetus. The blastocyst sticks securely to the lining, where it receives nourishment via the motherís bloodstream.
The cells of the embryo, an organism in its earliest stage of development, continue to multiply and begin to take on specific functions. A major portion of DNA is involved in regulating genetic functions. A process called differentiation leads to the various cell types that make up a human being. There is rapid growth, and the babyís organs and principal external features begin to take form. The period between conception and birth, during which the fetus grows and matures inside the motherís womb, is called gestation. In humans, the length of pregnancy is about nine months.
Diseases
Humans share the world with a multitude of plants and animals that are easily seen and recognized, and also with a vast assortment of microorganisms, unseen and unknown until the nineteenth century.
Diseases are abnormal health conditions that cause suffering, pain, functional failings or death. They may be caused by infections or by internal causes such as degenerative and autoimmune diseases.
Infectious diseases and the type of agents that cause them include tuberculosis (mycobacteria), malaria (Plasmodium protozoa), influenza (influenza virus), and blastomycosis (Blastomyces fungi). Infectious worms such as pinworms and tapeworms are parasites that use humans as hosts. The human body incorporates a powerful immune system that functions to mitigate or suppress infectious diseases. Immune system antibodies and T cells identify and respond to invasive microorganisms, often destroying them or rendering them inactive before they can cause a disease.
Autoimmune diseases are caused by abnormal immune system responses and are often associated with DNA irregularities. For example, psoriasis is linked to an allele of the gene PSORS1 in chromosome 6, and diabetes mellitus type 1 is linked with an allele of gene IDDM1 in chromosome 6.
Genetic diseases are caused by genome abnormalities or maladaptations, often linked to mutations in specific genes, or by errors in reproductive cell division. For example, cystic fibrosis is caused by a mutation in the CFTR gene in chromosome 7, and Down syndrome is caused by the presence of an extra copy of chromosome 21 in an individualís genome.
Ageing is among the most important risk factors for human diseases. American physician and microbiologist Leonard Hayflick discovered in 1962 that cultured normal human and animal cells have a limited capacity for replication. The human cells in Hayflickís experiment stopped replicating after about 60 mitosis cycles. Discovery of the Hayflick Limit countered the notion that normal cells would proliferate indefinitely in culture. It is currently thought that for vertebrates only tumor cells are immortal. An organism with normal cells that cannot replicate forever will eventually die. For humans, the upper lifespan limit is currently about 130 years. Age-related diseases include degenerative diseases such as heart disease, osteoporosis, prostatitis, arthritis, and dementia.
Non-Mendelian Inheritance
The mechanism of Mendelian inheritance has been confirmed by many genetic studies, but experiments in artificial selection and other studies have also identified instances of non-Mendelian inheritance in humans and other organisms.Mendel noted a characteristic in his peas that combined brown seed coat, violet flowers, and axial spots. In 1910, German zoologist Ludwig Plate used the term pleiotropy to refer to instances where one gene influences multiple traits. In contrast, other genetic studies led to the development of the concept of polygenic inheritance, for traits that are influenced by multiple genes.
The underlying mechanism in pleiotropy is a gene that codes for a product used by various cell types, or has an influence on various targets. In polygenic inheritance, a trait, for example, height in humans, is influenced by various DNA loci, with each DNA locus separately subject to inheritance. Pleiotropy and polygenic inheritance are examples of non-Mendelian inheritance.
In 1901, Karl Landsteiner, working at the University of Vienna, identified four principal blood types in humans. Compatibility of blood types is important because it can make the difference between a blood transfusion being beneficial and being harmful. Ten years later, Polish microbiologist Ludwik Hirszfeld and German physiologist Emil von Dungern, working at the Heidelberg Institute for Experimental Cancer Research in Germany, showed that the principal blood types A, B, AB, and O, are inherited.
An individualís blood type is determined from the inheritance of one allele from each parent. There are three different alleles: A, B, and O, which can result in six distinct combinations (AA, AB, AO, BB, BO, and OO). The alleles A and B are both dominant and the allele O is recessive, so the AO and BO genotypes result in A and B phenotypes, respectively. The occurrence of the genotype AB results in both alleles A and B being expressed (the alleles are codominant). Individuals with blood type AB show the characteristics of both type A and type B blood.
| Genotype | Phenotype |
| AA or AO | A |
| BB or BO | B |
| AB | A, B |
| OO | O |
ABO blood type inheritance, where a trait is determined by multiple alleles, is a form of inheritance which does not follow the Mendelian model of a single pair of alleles.
In 1923, John Gerould, then with Dartmouth College, published Inheritance of White Wing Color, a Sex-Limited Variation in Yellow Pierid Butterflies. Gerouldís experiments with butterflies of the genus Colias indicated that certain alleles were inherited by both males and females, but the associated characteristic was only expressed in one sex. In this type of inheritance, sex limits the expression of traits not obviously connected with reproduction. In humans, an example of this is facial hair, which is present in both men and women, but fully expressed as a heavy beard only in males.
The snapdragon plant (genus Antirrhinum) is subject to a type of inheritance in which there is incomplete dominance of a dominant trait over the corresponding recessive trait. This is observed in the inheritance of the snapdragon flower color characteristic. The red color trait is dominant (A) and the white color (a) is recessive. Inheritance of the homozygous dominant allele AA results in a red flower, and inheritance of the homozygous recessive allele aa results in a white flower, as predicted by Mendelís First Law, but inheritance of the heterozygous allele Aa does not result in a red flower. Instead, for snapdragon flowers, inheritance of the allele Aa results in an intermediate phenotype with pink color. This is an example of incomplete dominance by the dominant trait.
In certain insect species, males and females are quite different genetically. Ants, of the family Formicidae, and their related species, bees and wasps, have no sex chromosomes. Female ants have two copies of the genome and males have only one. In these species, the whole genome can function like a sex chromosome. A gene's effect may be masked or canceled by a different version of the same gene. In this manner, a gene can be harmful and recessive in females but favorable and dominant in males, where it can't be masked by a second copy.
Genome Comparisons
Hundreds of genomes from different organisms have been sequenced. The following table presents information comparing genome characteristics of organisms from selected species.
| Comparison of Genome Characteristics | ||||
| Organism | Type | Genome Size (base pairs) | Number of Chromosomes | Number of Genes |
| Phage Phi-X174 | Virus | 5.4 kB | 1 | 11 |
| Haemophilius influenzae | Bacterium | 1.8 MB | 1 | 1,801 |
| Saccharomyces cerevisiae (yeast) | Fungus | 12.1 MB | 32 | 6,275 |
| Arabidopsis thaliana | Plant | 135 MB | 10 | 27,655 |
| Quercus gilva (red-bark oak) | Plant | 890 MB | 12 | 36,442 |
| Populus trichocarpa (black cottonwood) | Plant | 480 MB | 38 | 73,013 |
| Caenorhabditis elegans (roundworm) | Nematode | 100 MB | 12 | 19,000 |
| Drosophila melanogaster (fruit fly) | Insect | 139.5 MB | 8 | 15,682 |
| Apis mellifera (honey bee) | Insect | 236 MB | 32 fem, 16 male | 10,157 |
| Pantala flavescens (skimmer dragonfly) | Insect | 633.3 MB | 12 | 15,354 |
| Platyplectrum ornatum (ornate burrowing frog) | Amphibian | 1.06 GB | 22 | 21,913 |
| Pleurodeles waltl (Iberian ribbed newt) | Amphibian | 19.4 GB | 24 | 22,847 |
| Tetraodon nigroviridis (puffer fish) | Fish | 390 MB | 42 | 22,400 |
| Gallus gallus (chicken) | Bird | 1.0 GB | 78 | 20,000 |
| Mus musculus (mouse) | Mammal | 2.7 GB | 40 | 20,210 |
| Felis silvestris catus (cat) | Mammal | 2.35 GB | 38 | 21,348 |
| Equus caballus (horse) | Mammal | 2.41 GB | 32 | 21,468 |
| Nomascus leucogenys (white-cheeked gibbon) | Mammal | 2.9 GB | 52 | 18,575 |
| Pan troglodytes (chimpanzee) | Mammal | 3.3 GB | 48 | 20,000 |
| Homo sapiens (human) | Mammal | 3.1 GB | 46 | 20,000 |
|